Mushroom development
Project description
Mushrooms are the reproductive structures of certain species of fungi, and they are a nutritious and sustainable food source for a growing world population. Very little is known about how mushrooms form. For example, only a handful of genes are known to be involved in this developmental process. The aim of this project is to gain more insight in the regulation of mushroom development. Specifically, we want to know which regulatory genes (transcription factors) control the initiation of mushroom growth and development. More knowledge about mushroom development will eventually lead to more efficient cultivation of additional species of mushroom-forming fungi.
Most mushroom-forming fungi are very difficult to work with in the lab, since they grow very slowly or because they are not genetically accessible. For this reason we use the model mushroom Schizophyllum commune . We have sequenced the genomes of several interesting strains of this species, and we determined how genes are regulated during mushroom development. Importantly, we have recently developed a technique based on CRISPR/Cas9 to efficiently make edits to the genome, allowing us to study gene function with much greater ease. We currently use this technique (as well as other approaches) to study the role of important regulatory genes during mushroom development.
The project is funded with the ERC Starting Grant awarded to Robin Ohm.
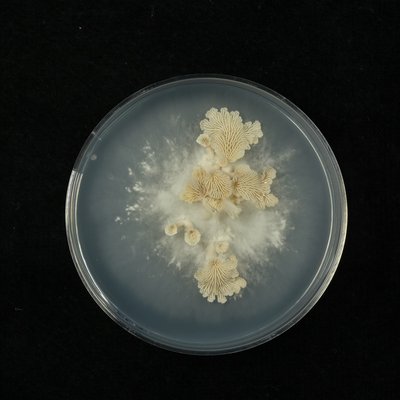
Figure. Mushroom development in the model system Schizophyllum commune.
Lab members currently working on this project
- Robin Ohm (Assistant professor)
Genome portals
- Agaricus bisporus var bisporus H39
- Agaricus bisporus var bisporus H97
- Agaricus bisporus var burnettii H119p4
Publications
High phenotypic and genotypic plasticity among strains of the mushroom-forming fungus Schizophyllum commune.Marian IM, Valdes ID, Hayes RD, LaButti K, Duffy K, Chovatia M, Johnson J, Ng V, Lugones LG, Wosten HAB, Grigoriev IV, Ohm RAFungal Genet Biol. 2024 Aug; 173: 103913. doi: 10.1016/j.fgb.2024.103913
The regulatory network of the White Collar complex during early mushroom development in Schizophyllum commune.Vonk PJ, van der Poel MJP, Niemeijer ZE, Ohm RAMicrobiol Res. 2024 Jul; 284: 127736. doi: 10.1016/j.micres.2024.127736
Lessons on fruiting body morphogenesis from genomes and transcriptomes of Agaricomycetes.Nagy LG, Vonk PJ, Kunzler M, Foldi C, Viragh M, Ohm RA, Hennicke F, Balint B, Csernetics A, Hegedus B, Hou Z, Liu XB, Nan S, Pareek M, Sahu N, Szathmari B, Varga T, Wu H, Yang X, Merenyi ZStud Mycol. 2023 Jul; 104: 1-85. doi: 10.3114/sim.2022.104.01
H3K4me2 ChIP-Seq reveals the epigenetic landscape during mushroom formation and novel developmental regulators of Schizophyllum commune.Vonk PJ, Ohm RASci Rep. 2021 Apr; 11(1): 8178. doi: 10.1038/s41598-021-87635-8
Telomere-to-telomere assembled and centromere annotated genomes of the two main subspecies of the button mushroom Agaricus bisporus reveal especially polymorphic chromosome ends.Sonnenberg ASM, Sedaghat-Telgerd N, Lavrijssen B, Ohm RA, Hendrickx PM, Scholtmeijer K, Baars JJP, van Peer ASci Rep. 2020 Sep; 10(1): 14653. doi: 10.1038/s41598-020-71043-5
High-throughput targeted gene deletion in the model mushroom Schizophyllum commune using pre-assembled Cas9 ribonucleoproteins.Vonk PJ, Escobar N, Wosten HAB, Lugones LG, Ohm RASci Rep. 2019 May; 9(1): 7632. doi: 10.1038/s41598-019-44133-2
The role of homeodomain transcription factors in fungal developmentVonk PJ, Ohm RAFungal Biol Rev. 2018 Sep; 34(4): 219-230. doi: 10.1016/j.fbr.2018.04.002
The blue light receptor complex WC-1/2 of Schizophyllum commune is involved in mushroom formation and protection against phototoxicity.Ohm RA, Aerts D, Wosten HA, Lugones LGEnviron Microbiol. 2013 Mar; 15(3): 943-55. doi: 10.1111/j.1462-2920.2012.02878.x
Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation.Ohm RA, de Jong JF, de Bekker C, Wosten HA, Lugones LGMol Microbiol. 2011 Sep; 81(6): 1433-45. doi: 10.1111/j.1365-2958.2011.07776.x
An efficient gene deletion procedure for the mushroom-forming basidiomycete Schizophyllum commune.Ohm RA, de Jong JF, Berends E, Wang F, Wosten HA, Lugones LGWorld J Microbiol Biotechnol. 2010 Oct; 26(10): 1919-1923. doi: 10.1007/s11274-010-0356-0
Genome sequence of the model mushroom Schizophyllum commune.Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, de Vries RP, Record E, Levasseur A, Baker SE, Bartholomew KA, Coutinho PM, Erdmann S, Fowler TJ, Gathman AC, Lombard V, Henrissat B, Knabe N, Kues U, Lilly WW, Lindquist E, Lucas S, Magnuson JK, Piumi F, Raudaskoski M, Salamov A, Schmutz J, Schwarze FW, vanKuyk PA, Horton JS, Grigoriev IV, Wosten HANat Biotechnol. 2010 Sep; 28(9): 957-63. doi: 10.1038/nbt.1643
Inactivation of ku80 in the mushroom-forming fungus Schizophyllum commune increases the relative incidence of homologous recombination.de Jong JF, Ohm RA, de Bekker C, Wosten HA, Lugones LGFEMS Microbiol Lett. 2010 Sep; 310(1): 91-5. doi: 10.1111/j.1574-6968.2010.02052.x
