Fungal comparative genomics
Project description
In the past decade, the number of available fungal genome sequences has increased considerably. Moreover, it has become much cheaper to generate high-quality sequences of fungal genomes.
In this on-going project, we analyze fungal genomes (newly sequenced and previously published) to gain more insight into the fungal lifestyle and development. This is done both in collaboration with other labs (including the Joint Genome Institute), and by sequencing new genomes ourselves.
Furthermore, we developed Genome Portals that will facilitate the analysis of the genomes we have sequenced in-house.
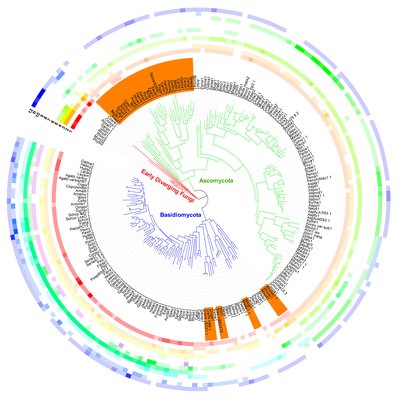
Figure. Homeodomain transcription factors in the fungal kingdom. (Vonk et al., Fungal Biology Reviews, 2018)
Lab members currently working on this project
- Robin Ohm (Assistant professor)
Genome portals
- Agaricus bisporus var bisporus H39
- Agaricus bisporus var bisporus H97
- Agaricus bisporus var burnettii H119p4
Publications
High phenotypic and genotypic plasticity among strains of the mushroom-forming fungus Schizophyllum commune.Marian IM, Valdes ID, Hayes RD, LaButti K, Duffy K, Chovatia M, Johnson J, Ng V, Lugones LG, Wosten HAB, Grigoriev IV, Ohm RAFungal Genet Biol. 2024 Aug; 173: 103913. doi: 10.1016/j.fgb.2024.103913
Genome annotations for the ascomycete fungi Trichoderma harzianum, Trichoderma aggressivum, and Purpureocillium lilacinum.Beijen EPW, Ohm RAMicrobiol Resour Announc. 2024 Mar; 13(3): e0115323. doi: 10.1128/mra.01153-23
Genome-scale phylogeny and comparative genomics of the fungal order Sordariales.Hensen N, Bonometti L, Westerberg I, Brannstrom IO, Guillou S, Cros-Aarteil S, Calhoun S, Haridas S, Kuo A, Mondo S, Pangilinan J, Riley R, LaButti K, Andreopoulos B, Lipzen A, Chen C, Yan M, Daum C, Ng V, Clum A, Steindorff A, Ohm RA, Martin F, Silar P, Natvig DO, Lalanne C, Gautier V, Ament-Velasquez SL, Kruys A, Hutchinson MI, Powell AJ, Barry K, Miller AN, Grigoriev IV, Debuchy R, Gladieux P, Hiltunen Thoren M, Johannesson HMol Phylogenet Evol. 2023 Dec; 189: 107938. doi: 10.1016/j.ympev.2023.107938
Genome sequences of 24 Aspergillus niger sensu stricto strains to study strain diversity, heterokaryon compatibility, and sexual reproduction.Seekles SJ, Punt M, Savelkoel N, Houbraken J, Wosten HAB, Ohm RA, Ram AFJG3 (Bethesda). 2022 Jul; 12(7). doi: 10.1093/g3journal/jkac124
Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits.Miyauchi S, Kiss E, Kuo A, Drula E, Kohler A, Sanchez-Garcia M, Morin E, Andreopoulos B, Barry KW, Bonito G, Buee M, Carver A, Chen C, Cichocki N, Clum A, Culley D, Crous PW, Fauchery L, Girlanda M, Hayes RD, Keri Z, LaButti K, Lipzen A, Lombard V, Magnuson J, Maillard F, Murat C, Nolan M, Ohm RA, Pangilinan J, Pereira MF, Perotto S, Peter M, Pfister S, Riley R, Sitrit Y, Stielow JB, Szollosi G, Zifcakova L, Stursova M, Spatafora JW, Tedersoo L, Vaario LM, Yamada A, Yan M, Wang P, Xu J, Bruns T, Baldrian P, Vilgalys R, Dunand C, Henrissat B, Grigoriev IV, Hibbett D, Nagy LG, Martin FMNat Commun. 2020 Oct; 11(1): 5125. doi: 10.1038/s41467-020-18795-w
Degradative Capacity of Two Strains of Rhodonia placenta: From Phenotype to Genotype.Kolle M, Horta MAC, Nowrousian M, Ohm RA, Benz JP, Pilgard AFront Microbiol. 2020 Jul; 11: 1338. doi: 10.3389/fmicb.2020.01338
101 Dothideomycetes genomes: A test case for predicting lifestyles and emergence of pathogens.Haridas S, Albert R, Binder M, Bloem J, LaButti K, Salamov A, Andreopoulos B, Baker SE, Barry K, Bills G, Bluhm BH, Cannon C, Castanera R, Culley DE, Daum C, Ezra D, Gonzalez JB, Henrissat B, Kuo A, Liang C, Lipzen A, Lutzoni F, Magnuson J, Mondo SJ, Nolan M, Ohm RA, Pangilinan J, Park HJ, Ramirez L, Alfaro M, Sun H, Tritt A, Yoshinaga Y, Zwiers LH, Turgeon BG, Goodwin SB, Spatafora JW, Crous PW, Grigoriev IVStud Mycol. 2020 Jun; 96: 141-153. doi: 10.1016/j.simyco.2020.01.003
Megaphylogeny resolves global patterns of mushroom evolution.Varga T, Krizsan K, Foldi C, Dima B, Sanchez-Garcia M, Sanchez-Ramirez S, Szollosi GJ, Szarkandi JG, Papp V, Albert L, Andreopoulos W, Angelini C, Antonin V, Barry KW, Bougher NL, Buchanan P, Buyck B, Bense V, Catcheside P, Chovatia M, Cooper J, Damon W, Desjardin D, Finy P, Geml J, Haridas S, Hughes K, Justo A, Karasinski D, Kautmanova I, Kiss B, Kocsube S, Kotiranta H, LaButti KM, Lechner BE, Liimatainen K, Lipzen A, Lukacs Z, Mihaltcheva S, Morgado LN, Niskanen T, Noordeloos ME, Ohm RA, Ortiz-Santana B, Ovrebo C, Racz N, Riley R, Savchenko A, Shiryaev A, Soop K, Spirin V, Szebenyi C, Tomsovsky M, Tulloss RE, Uehling J, Grigoriev IV, Vagvolgyi C, Papp T, Martin FM, Miettinen O, Hibbett DS, Nagy LGNat Ecol Evol. 2019 Apr; 3(4): 668-678. doi: 10.1038/s41559-019-0834-1
Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi.Krizsan K, Almasi E, Merenyi Z, Sahu N, Viragh M, Koszo T, Mondo S, Kiss B, Balint B, Kues U, Barry K, Cseklye J, Hegedus B, Henrissat B, Johnson J, Lipzen A, Ohm RA, Nagy I, Pangilinan J, Yan J, Xiong Y, Grigoriev IV, Hibbett DS, Nagy LGProc Natl Acad Sci U S A. 2019 Apr; 116(15): 7409-7418. doi: 10.1073/pnas.1817822116
The role of homeodomain transcription factors in fungal developmentVonk PJ, Ohm RAFungal Biol Rev. 2018 Sep; 34(4): 219-230. doi: 10.1016/j.fbr.2018.04.002
Genome-Wide Analysis of Corynespora cassiicola Leaf Fall Disease Putative Effectors.Lopez D, Ribeiro S, Label P, Fumanal B, Venisse JS, Kohler A, de Oliveira RR, Labutti K, Lipzen A, Lail K, Bauer D, Ohm RA, Barry KW, Spatafora J, Grigoriev IV, Martin FM, Pujade-Renaud VFront Microbiol. 2018 Jun; 9: 276. doi: 10.3389/fmicb.2018.00276
Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi.Knapp DG, Nemeth JB, Barry K, Hainaut M, Henrissat B, Johnson J, Kuo A, Lim JHP, Lipzen A, Nolan M, Ohm RA, Tamas L, Grigoriev IV, Spatafora JW, Nagy LG, Kovacs GMSci Rep. 2018 Apr; 8(1): 6321. doi: 10.1038/s41598-018-24686-4
Comparative genomics and transcriptomics depict ericoid mycorrhizal fungi as versatile saprotrophs and plant mutualists.Martino E, Morin E, Grelet GA, Kuo A, Kohler A, Daghino S, Barry KW, Cichocki N, Clum A, Dockter RB, Hainaut M, Kuo RC, LaButti K, Lindahl BD, Lindquist EA, Lipzen A, Khouja HR, Magnuson J, Murat C, Ohm RA, Singer SW, Spatafora JW, Wang M, Veneault-Fourrey C, Henrissat B, Grigoriev IV, Martin FM, Perotto SNew Phytol. 2018 Feb; 217(3): 1213-1229. doi: 10.1111/nph.14974
Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens.Uehling J, Gryganskyi A, Hameed K, Tschaplinski T, Misztal PK, Wu S, Desiro A, Vande Pol N, Du Z, Zienkiewicz A, Zienkiewicz K, Morin E, Tisserant E, Splivallo R, Hainaut M, Henrissat B, Ohm R, Kuo A, Yan J, Lipzen A, Nolan M, LaButti K, Barry K, Goldstein AH, Labbe J, Schadt C, Tuskan G, Grigoriev I, Martin F, Vilgalys R, Bonito GEnviron Microbiol. 2017 Aug; 19(8): 2964-2983. doi: 10.1111/1462-2920.13669
Ectomycorrhizal ecology is imprinted in the genome of the dominant symbiotic fungus Cenococcum geophilum.Peter M, Kohler A, Ohm RA, Kuo A, Krutzmann J, Morin E, Arend M, Barry KW, Binder M, Choi C, Clum A, Copeland A, Grisel N, Haridas S, Kipfer T, LaButti K, Lindquist E, Lipzen A, Maire R, Meier B, Mihaltcheva S, Molinier V, Murat C, Poggeler S, Quandt CA, Sperisen C, Tritt A, Tisserant E, Crous PW, Henrissat B, Nehls U, Egli S, Spatafora JW, Grigoriev IV, Martin FMNat Commun. 2016 Sep; 7: 12662. doi: 10.1038/ncomms12662
Expansion of Signal Transduction Pathways in Fungi by Extensive Genome Duplication.Corrochano LM, Kuo A, Marcet-Houben M, Polaino S, Salamov A, Villalobos-Escobedo JM, Grimwood J, Alvarez MI, Avalos J, Bauer D, Benito EP, Benoit I, Burger G, Camino LP, Canovas D, Cerda-Olmedo E, Cheng JF, Dominguez A, Elias M, Eslava AP, Glaser F, Gutierrez G, Heitman J, Henrissat B, Iturriaga EA, Lang BF, Lavin JL, Lee SC, Li W, Lindquist E, Lopez-Garcia S, Luque EM, Marcos AT, Martin J, McCluskey K, Medina HR, Miralles-Duran A, Miyazaki A, Munoz-Torres E, Oguiza JA, Ohm RA, Olmedo M, Orejas M, Ortiz-Castellanos L, Pisabarro AG, Rodriguez-Romero J, Ruiz-Herrera J, Ruiz-Vazquez R, Sanz C, Schackwitz W, Shahriari M, Shelest E, Silva-Franco F, Soanes D, Syed K, Tagua VG, Talbot NJ, Thon MR, Tice H, de Vries RP, Wiebenga A, Yadav JS, Braun EL, Baker SE, Garre V, Schmutz J, Horwitz BA, Torres-Martinez S, Idnurm A, Herrera-Estrella A, Gabaldon T, Grigoriev IVCurr Biol. 2016 Jun; 26(12): 1577-1584. doi: 10.1016/j.cub.2016.04.038
Comparative Genomics of Early-Diverging Mushroom-Forming Fungi Provides Insights into the Origins of Lignocellulose Decay Capabilities.Nagy LG, Riley R, Tritt A, Adam C, Daum C, Floudas D, Sun H, Yadav JS, Pangilinan J, Larsson KH, Matsuura K, Barry K, Labutti K, Kuo R, Ohm RA, Bhattacharya SS, Shirouzu T, Yoshinaga Y, Martin FM, Grigoriev IV, Hibbett DSMol Biol Evol. 2016 Apr; 33(4): 959-70. doi: 10.1093/molbev/msv337
Genomics of wood-degrading fungi.Ohm RA, Riley R, Salamov A, Min B, Choi IG, Grigoriev IVFungal Genet Biol. 2014 Nov; 72: 82-90. doi: 10.1016/j.fgb.2014.05.001
Prevalence of transcription factors in ascomycete and basidiomycete fungi.Todd RB, Zhou M, Ohm RA, Leeggangers HA, Visser L, de Vries RPBMC Genomics. 2014 Mar; 15: 214. doi: 10.1186/1471-2164-15-214
Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi.Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, Barry KW, Condon BJ, Copeland AC, Dhillon B, Glaser F, Hesse CN, Kosti I, LaButti K, Lindquist EA, Lucas S, Salamov AA, Bradshaw RE, Ciuffetti L, Hamelin RC, Kema GH, Lawrence C, Scott JA, Spatafora JW, Turgeon BG, de Wit PJ, Zhong S, Goodwin SB, Grigoriev IVPLoS Pathog. 2012 Jun; 8(12): e1003037. doi: 10.1371/journal.ppat.1003037
