Defense of mushrooms against their fungal competitors
Project description
Mushrooms are a nutritious and sustainable food source for a growing world population. However, mushroom-forming fungi are prone to infections by a range of fungal and bacterial competitors, which can lead to devastating crop losses. A solution is to develop strains that are more resistant, but this requires understanding of the fungal innate immune system. Relatively little is currently known about this, especially compared to animals and plants. No regulatory components of the defense against pathogens have been identified in mushroom-forming fungi. The aim of this project is to identify the gene regulatory network and signal transduction pathways involved in defense of mushroom-forming fungi against their (fungal) competitors.
This project is funded by a VIDI grant from NWO (Dutch Research Council) awarded to Robin Ohm.
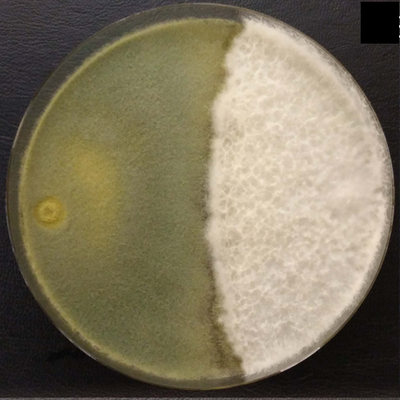
Figure. The mushroom-forming fungus Schizophyllum commune (right, white mycelium) and its competitor Trichoderma harzianum (left, green sporulating mycelium).
Lab members currently working on this project
- Robin Ohm (Assistant professor)
- Erik Beijen (PhD student)
- Marieke van Maanen (PhD student)
- Gaya Verhagen (Master student)
- Laura Knittel (Master student)
- Sofia Saveedra Ravest (Master student)
Genome portals
- Purpureocillium lilacinum CBS 150709
- Trichoderma aggressivum f. europaeum CBS 100526
- Trichoderma harzianum CBS 354.33
Publications
Transcriptomics reveals the regulation of the immune system of the mushroom-forming fungus Schizophyllum commune during interaction with four competitors.Beijen EPW, van Maanen MH, Marian IM, Klusener JX, van Roosmalen E, Herman KC, Koster MC, Ohm RAMicrobiol Res. 2024 Dec; 289: 127929. doi: 10.1016/j.micres.2024.127929
Genome annotations for the ascomycete fungi Trichoderma harzianum, Trichoderma aggressivum, and Purpureocillium lilacinum.Beijen EPW, Ohm RAMicrobiol Resour Announc. 2024 Mar; 13(3): e0115323. doi: 10.1128/mra.01153-23
